The Migale bioinformatics facility give acces to its users to a R environment (version 4.02) with more than 1000 packages installed.
For your development and prototyping activities with R, we now give you access to the RStudio development environment through a web interface at this adress :
https://rstudio.migale.inrae.fr
This RStudio is configured to use the R from Migale. It is linked to your work directories on the platform.
 Migale Bioinformatics Facility
Migale Bioinformatics Facility
We provide several services for scientists to deal with life sciences data
Our Services
Migale, one of the Collective Scientific Infrastructure of INRAE, is part of the BioinfOmics Research Infrastructure of INRAE for bioinformatics. It is also a member of IFB (Institut Français de Bioinformatique), the French bioinformatics infrastructure and associated facility of France Génomique, the French genomic infrastructure for which we contribute to support different developments in bioinformatics.
A free account gives you access to work and save directories for your data, and access to the computer farm for your analyses.
The cluster farm is composed of about a thousand cores organized in different queues. We use the Sun Grid Engine queuing system for managing jobs.
You have a free access to our Galaxy server. Galaxy allows non-bioinformaticians to easily run tools without technical knowledges.
Command line tools, R packages and Galaxy wrappers are available on request and accessible to all migale authenticated users.
We provide an access to a large set of public biological databanks including whole genomes, nucleic and proteic sequences and other resources. They are updated automatically with BioMaJ or upon request.
We write tutorials to help you get familiar with tools, best practices, languages, etc.
Each year, we offer our "Bioinformatics by practicing" cycle. This cycle covers a broad spectrum of bioinformatics. The modules mix theoretical part and practical work.
We answer to the most common questions regarding the technical difficulties you can go through on our infrastructure.
Find all the ways to contact us.
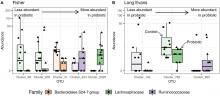
New publication: A Probiotic Mixture Induces Anxiolytic- and Antidepressive-Like Effects in Fischer and Maternally Deprived Long Evans Rats
Mahendra and Olivier were sollicited by Valérie Daugé at MICALIS institute to analyze feces microbiota (V3-V4 regions of the 16S rDNA gene): α- and β-diversities and differential abundance analyses of OTUs.
Biological results:
Update R
R 4.0.2 is now the default version on migale
CHANGES IN R 4.0.2
UTILITIES:
* R CMD check skips vignette re-building (with a warning) if the VignetteBuilder package(s) are not available.
BUG FIXES:
* Paths with non-ASCII characters caused problems for package loading on Windows PR#17833.
* Using tcltk widgets no longer crashes R on Windows.
* source(*, echo=TRUE) no longer fails in some cases with empty lines; reported by Bill Dunlap in PR#17769.
* on.exit() now correctly matches named arguments, thanks to PR#17815